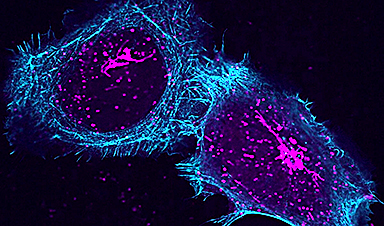
Researchers are constantly expanding their arsenal of methods to decipher the spatial organization of biological structures. Using microscopes, they can now visualize individual macromolecular components within DNA, protein, or other complexes. However, this resolution typically requires sophisticated equipment applied to specially-processed samples, and it is difficult to simultaneously watch many types of molecules, especially at high density and throughput, or dynamic interactions.
Circumventing the need for expensive microscopes, some recent biochemical approaches attach barcoded DNA probes to molecular targets and then fuse those in nearby pairs together, often by DNA ligation. These DNA “records” are later read out for analysis.
Because these methods destroy the DNA probes in the process of pairing, however, the information acquired from each molecular target cannot include more than one interaction, neither multiple at once nor one changing over time. Such methods can severely limit the quality of any subsequent computational reconstruction, and make reconstruction of individual complexes impossible.
To overcome these limitations, a team at Harvard’s Wyss Institute of Biologically Inspired Engineering led by Core Faculty member Peng Yin, Ph.D., has now developed a DNA nanotechnology-based method that allows for repeated, non-destructive recording of uniquely barcoded molecular pairings, rendering a detailed view of their components and geometries.
Image Credit: vimeo.com
Recent News
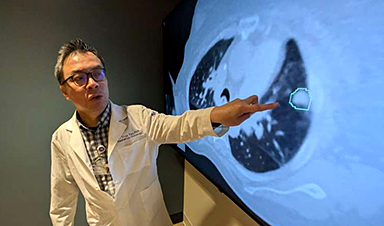
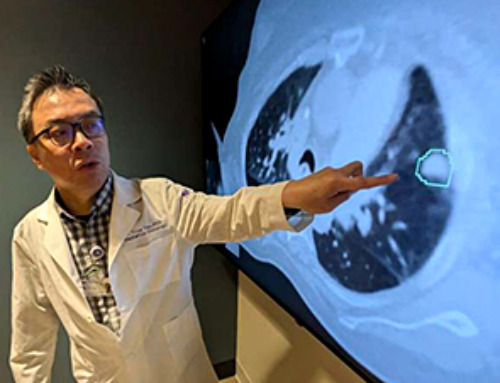
AI matches doctors in mapping lung tumors for radiation therapy
In radiation therapy, precision can save lives. Oncologists must carefully map the size and location of a tumor before delivering high-dose radiation to destroy cancer cells while sparing healthy [...]
Scientists Finally “See” Key Protein That Controls Inflammation
Researchers used advanced microscopy to uncover important protein structures. For the first time, two important protein structures in the human body are being visualized, thanks in part to cutting-edge [...]
AI tool detects 9 types of dementia from a single brain scan
Mayo Clinic researchers have developed a new artificial intelligence (AI) tool that helps clinicians identify brain activity patterns linked to nine types of dementia, including Alzheimer's disease, using a [...]
Is plastic packaging putting more than just food on your plate?
New research reveals that common food packaging and utensils can shed microscopic plastics into our food, prompting urgent calls for stricter testing and updated regulations to protect public health. [...]
Aging Spreads Through the Bloodstream
Summary: New research reveals that aging isn’t just a local cellular process—it can spread throughout the body via the bloodstream. A redox-sensitive protein called ReHMGB1, secreted by senescent cells, was [...]
AI and nanomedicine find rare biomarkers for prostrate cancer and atherosclerosis
Imagine a stadium packed with 75,000 fans, all wearing green and white jerseys—except one person in a solid green shirt. Finding that person would be tough. That's how hard [...]
Are Pesticides Breeding the Next Pandemic? Experts Warn of Fungal Superbugs
Fungicides used in agriculture have been linked to an increase in resistance to antifungal drugs in both humans and animals. Fungal infections are on the rise, and two UC Davis infectious [...]
Scientists Crack the 500-Million-Year-Old Code That Controls Your Immune System
A collaborative team from Penn Medicine and Penn Engineering has uncovered the mathematical principles behind a 500-million-year-old protein network that determines whether foreign materials are recognized as friend or [...]
Team discovers how tiny parts of cells stay organized, new insights for blocking cancer growth
A team of international researchers led by scientists at City of Hope provides the most thorough account yet of an elusive target for cancer treatment. Published in Science Advances, the [...]
Nanomaterials in Ophthalmology: A Review
Eye diseases are becoming more common. In 2020, over 250 million people had mild vision problems, and 295 million experienced moderate to severe ocular conditions. In response, researchers are [...]
Natural Plant Extract Removes up to 90% of Microplastics From Water
Researchers found that natural polymers derived from okra and fenugreek are highly effective at removing microplastics from water. The same sticky substances that make okra slimy and give fenugreek [...]
Instant coffee may damage your eyes, genetic study finds
A new genetic study shows that just one extra cup of instant coffee a day could significantly increase your risk of developing dry AMD, shedding fresh light on how [...]
Nanoneedle patch offers painless alternative to traditional cancer biopsies
A patch containing tens of millions of microscopic nanoneedles could soon replace traditional biopsies, scientists have found. The patch offers a painless and less invasive alternative for millions of [...]
Small antibodies provide broad protection against SARS coronaviruses
Scientists have discovered a unique class of small antibodies that are strongly protective against a wide range of SARS coronaviruses, including SARS-CoV-1 and numerous early and recent SARS-CoV-2 variants. [...]
Controlling This One Molecule Could Halt Alzheimer’s in Its Tracks
New research identifies the immune molecule STING as a driver of brain damage in Alzheimer’s. A new approach to Alzheimer’s disease has led to an exciting discovery that could help [...]
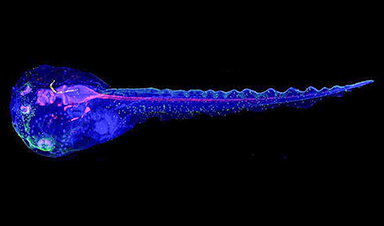
Cyborg tadpoles are helping us learn how brain development starts
How does our brain, which is capable of generating complex thoughts, actions and even self-reflection, grow out of essentially nothing? An experiment in tadpoles, in which an electronic implant [...]
Prime Editing: The Next Frontier in Genetic Medicine
By Dr. Chinta SidharthanReviewed by Benedette Cuffari, M.Sc. Discover how prime editing is redefining the future of medicine by offering highly precise, safe, and versatile DNA corrections, bringing hope for more [...]
Can scientists predict life longevity from a drop of blood?
Discover how a new epigenetic clock measures how fast you are really aging from just a drop of blood or saliva. A recent study published in the journal Nature Aging constructed [...]
What is different about the NB.1.8.1 Covid variant?
For many of us, Covid-19 feels like a chapter we’ve closed – along with the days of PCR tests, mask mandates and daily case updates. But while life may [...]
Scientists discover single cell creatures can learn new behaviours
It was previously thought that learning behaviours only applied to animals with complex brain and nervous systems, but a new study has proven that this may also occur in individual cells. As a result, this [...]
Virus which ’causes multiple organ failure’ found at popular Spanish holiday destination
British tourists planning trips to Spain have been warned after a deadly virus that can cause multiple organ failure has been detected in the country. The Foreign Office issued the alert [...]
Urgent health warning as dangerous new Covid virus from China triggers US outbreak
A dangerous new Covid variant from China is surging in California, health officials warn. The California Department of Public Health warned this week the highly contagious NB.1.8.1 strain has been detected in [...]
How the evolution of a single gene allowed the plague to adapt, prolonging the pandemics
Scientists have documented the way a single gene in the bacterium that causes bubonic plague, Yersinia pestis, allowed it to survive hundreds of years by adjusting its virulence and [...]
Inhalable Nanovaccines: The Future of Needle-Free Immunization
The COVID-19 pandemic highlighted the need for adaptable and scalable vaccine technologies. While mRNA vaccines have improved disease prevention, most are delivered by intramuscular injection, which may not effectively [...]
‘Stealthy’ lipid nanoparticles give mRNA vaccines a makeover
A new material developed at Cornell University could significantly improve the delivery and effectiveness of mRNA vaccines by replacing a commonly used ingredient that may trigger unwanted immune responses [...]
You could be inhaling nearly 70,000 plastic particles annually, what it means for your health
Invisible plastics in the air are infiltrating our bodies and cities. Scientists reveal the urgent health dangers and outline bold solutions for a cleaner, safer future. In a recent [...]
Experts explain how H5 avian influenza adapts to infect more animals
A new global review reveals how rapidly evolving H5 bird flu viruses are reaching new species, including dairy cattle, and stresses the urgent need for coordinated action to prevent [...]
3D-printed device enables precise modeling of complex human tissues in the lab
A new, easily adopted, 3D-printed device will enable scientists to create models of human tissue with even greater control and complexity. An interdisciplinary group of researchers at the University [...]
Ancient DNA sheds light on evolution of relapsing fever bacteria
Researchers at the Francis Crick Institute and UCL have analyzed ancient DNA from Borrelia recurrentis, a type of bacteria that causes relapsing fever, pinpointing when it evolved to spread through [...]
Cold Sore Virus Linked to Alzheimer’s, Antivirals May Lower Risk
Summary: A large study suggests that symptomatic infection with herpes simplex virus 1 (HSV-1)—best known for causing cold sores—may significantly raise the risk of developing Alzheimer’s disease. Researchers found that [...]
Nanoparticle-Based Combination Therapy for Resistant Melanoma
A recent study published in Small addresses the persistent difficulty of treating refractory melanoma, an aggressive form of skin cancer that often does not respond to existing therapies. Although diagnostic tools [...]
Our DNA May Evolve Much Faster Than Previously Thought
Rapidly mutating DNA regions were mapped using a multi-generational family and advanced sequencing tools. Understanding how human DNA changes over generations is crucial for estimating genetic disease risks and tracing our [...]
AI therapy may help with mental health, but innovation should never outpace ethics
Mental health services around the world are stretched thinner than ever. Long wait times, barriers to accessing care and rising rates of depression and anxiety have made it harder for people to get timely help. As a result, governments [...]
Global life expectancy plunges as WHO warns of deepening health crisis Post-COVID
The World Health Organization (WHO) has sounded the alarm on the long-term health repercussions of the COVID-19 pandemic in its newly released World Health Statistics Report 2025. The report reveals [...]
Researchers map brain networks involved in word retrieval
How are we able to recall a word we want to say? This basic ability, called word retrieval, is often compromised in patients with brain damage. Interestingly, many patients [...]
Melting Ice Is Changing the Color of the Ocean – Scientists Are Alarmed
Melting sea ice changes not only how much light enters the ocean, but also its color, disrupting marine photosynthesis and altering Arctic ecosystems in subtle but profound ways. As global warming [...]
Your Washing Machine Might Be Helping Antibiotic-Resistant Bacteria Spread
A new study reveals that biofilms in washing machines may contain potential pathogens and antibiotic resistance genes, posing possible risks for laundering healthcare workers’ uniforms at home. Washing healthcare [...]
Scientists Discover Hidden Cause of Alzheimer’s Hiding in Plain Sight
Researchers found the PHGDH gene directly causes Alzheimer’s and discovered a drug-like molecule, NCT-503, that may help treat the disease early by targeting the gene’s hidden function. A recent study has [...]
How Brain Cells Talk: Inside the Complex Language of the Human Mind
Introduction The human brain contains nearly 86 billion neurons, constantly exchanging messages like an immense social media network, but neurons do not work alone – glial cells, neurotransmitters, receptors, and [...]
Oxford study reveals how COVID-19 vaccines prevent severe illness
A landmark study by scientists at the University of Oxford, has unveiled crucial insights into the way that COVID-19 vaccines mitigate severe illness in those who have been vaccinated. [...]
Annual blood test could detect cancer earlier and save lives
A single blood test, designed to pick up chemical signals indicative of the presence of many different types of cancer, could potentially thwart progression to advanced disease while the malignancy [...]
How the FDA opens the door to risky chemicals in America’s food supply
Lining the shelves of American supermarkets are food products with chemicals linked to health concerns. To a great extent, the FDA allows food companies to determine for themselves whether [...]
Superbug crisis could get worse, killing nearly 40 million people by 2050
The number of lives lost around the world due to infections that are resistant to the medications intended to treat them could increase nearly 70% by 2050, a new [...]
How Can Nanomaterials Be Programmed for Different Applications?
Nanomaterials are no longer just small—they are becoming smart. Across fields like medicine, electronics, energy, and materials science, researchers are now programming nanomaterials to behave in intentional, responsive ways. [...]
Microplastics Are Invading Our Arteries, and It Could Be Increasing Your Risk of Stroke
Higher levels of micronanoplastics were found in carotid artery plaque, especially in people with stroke symptoms, suggesting a potential new risk factor. People with plaque buildup in the arteries [...]
Gene-editing therapy shows early success in fighting advanced gastrointestinal cancers
Researchers at the University of Minnesota have completed a first-in-human clinical trial testing a CRISPR/Cas9 gene-editing technique to help the immune system fight advanced gastrointestinal (GI) cancers. The results, [...]
Engineered extracellular vesicles facilitate delivery of advanced medicines
Graphic abstract of the development of VEDIC and VFIC systems for high efficiency intracellular protein delivery in vitro and in vivo. Credit: Nature Communications (2025). DOI: 10.1038/s41467-025-59377-y. https://www.nature.com/articles/s41467-025-59377-y Researchers at Karolinska [...]
Brain-computer interface allows paralyzed users to customize their sense of touch
University of Pittsburgh School of Medicine scientists are one step closer to developing a brain-computer interface, or BCI, that allows people with tetraplegia to restore their lost sense of [...]
Scientists Flip a Gut Virus “Kill Switch” – Expose a Hidden Threat in Antibiotic Treatment
Scientists have long known that bacteriophages, viruses that infect bacteria, live in our gut, but exactly what they do has remained elusive. Researchers developed a clever mouse model that [...]
Enhanced Antibacterial Polylactic Acid-Curcumin Nanofibers for Wound Dressing
Background Wound healing is a complex physiological process that can be compromised by infection and impaired tissue regeneration. Conventional dressings, typically made from natural fibers such as cotton or [...]
Global Nanomaterial Regulation: A Country-by-Country Comparison
Nanomaterials are materials with at least one dimension smaller than 100 nanometres (about 100,000 times thinner than a human hair). Because of their tiny size, they have unique properties [...]
Pandemic Potential: Scientists Discover 3 Hotspots of Deadly Emerging Disease in the US
Virginia Tech researchers discovered six new rodent carriers of hantavirus and identified U.S. hotspots, highlighting the virus’s adaptability and the impact of climate and ecology on its spread. Hantavirus recently [...]
Studies detail high rates of long COVID among healthcare, dental workers
Researchers have estimated approximately 8% of Americas have ever experienced long COVID, or lasting symptoms, following an acute COVID-19 infection. Now two recent international studies suggest that the percentage is [...]
Melting Arctic Ice May Unleash Ancient Deadly Diseases, Scientists Warn
Melting Arctic ice increases human and animal interactions, raising the risk of infectious disease spread. Researchers urge early intervention and surveillance. Climate change is opening new pathways for the [...]
Scientists May Have Found a Secret Weapon To Stop Pancreatic Cancer Before It Starts
Researchers at Cold Spring Harbor Laboratory have found that blocking the FGFR2 and EGFR genes can stop early-stage pancreatic cancer from progressing, offering a promising path toward prevention. Pancreatic [...]
Breakthrough Drug Restores Vision: Researchers Successfully Reverse Retinal Damage
Blocking the PROX1 protein allowed KAIST researchers to regenerate damaged retinas and restore vision in mice. Vision is one of the most important human senses, yet more than 300 million people [...]
Differentiating cancerous and healthy cells through motion analysis
Researchers from Tokyo Metropolitan University have found that the motion of unlabeled cells can be used to tell whether they are cancerous or [...]
This Tiny Cellular Gate Could Be the Key to Curing Cancer – And Regrowing Hair
After more than five decades of mystery, scientists have finally unveiled the detailed structure and function of a long-theorized molecular machine in our mitochondria — the mitochondrial pyruvate carrier. [...]
Unlocking Vision’s Secrets: Researchers Reveal 3D Structure of Key Eye Protein
Researchers have uncovered the 3D structure of RBP3, a key protein in vision, revealing how it transports retinoids and fatty acids and how its dysfunction may lead to retinal [...]
5 Key Facts About Nanoplastics and How They Affect the Human Body
Nanoplastics are typically defined as plastic particles smaller than 1000 nanometers. These particles are increasingly being detected in human tissues: they can bypass biological barriers, accumulate in organs, and [...]
Measles Is Back: Doctors Warn of Dangerous Surge Across the U.S.
Parents are encouraged to contact their pediatrician if their child has been exposed to measles or is showing symptoms. Pediatric infectious disease experts are emphasizing the critical importance of [...]
AI at the Speed of Light: How Silicon Photonics Are Reinventing Hardware
A cutting-edge AI acceleration platform powered by light rather than electricity could revolutionize how AI is trained and deployed. Using photonic integrated circuits made from advanced III-V semiconductors, researchers have [...]
A Grain of Brain, 523 Million Synapses, Most Complicated Neuroscience Experiment Ever Attempted
A team of over 150 scientists has achieved what once seemed impossible: a complete wiring and activity map of a tiny section of a mammalian brain. This feat, part [...]
The Secret “Radar” Bacteria Use To Outsmart Their Enemies
A chemical radar allows bacteria to sense and eliminate predators. Investigating how microorganisms communicate deepens our understanding of the complex ecological interactions that shape our environment is an area [...]
Psychologists explore ethical issues associated with human-AI relationships
It's becoming increasingly commonplace for people to develop intimate, long-term relationships with artificial intelligence (AI) technologies. At their extreme, people have "married" their AI companions in non-legally binding ceremonies, [...]
When You Lose Weight, Where Does It Actually Go?
Most health professionals lack a clear understanding of how body fat is lost, often subscribing to misconceptions like fat converting to energy or muscle. The truth is, fat is [...]
How Everyday Plastics Quietly Turn Into DNA-Damaging Nanoparticles
The same unique structure that makes plastic so versatile also makes it susceptible to breaking down into harmful micro- and nanoscale particles. The world is saturated with trillions of microscopic and [...]
AI Outperforms Physicians in Real-World Urgent Care Decisions, Study Finds
The study, conducted at the virtual urgent care clinic Cedars-Sinai Connect in LA, compared recommendations given in about 500 visits of adult patients with relatively common symptoms – respiratory, [...]
Challenging the Big Bang: A Multi-Singularity Origin for the Universe
In a study published in the journal Classical and Quantum Gravity, Dr. Richard Lieu, a physics professor at The University of Alabama in Huntsville (UAH), which is a part of The University [...]
New drug restores vision by regenerating retinal nerves
Vision is one of the most crucial human senses, yet over 300 million people worldwide are at risk of vision loss due to various retinal diseases. While recent advancements [...]
Shingles vaccine cuts dementia risk by 20%, new study shows
A shingles shot may do more than prevent rash — it could help shield the aging brain from dementia, according to a landmark study using real-world data from the [...]
AI Predicts Sudden Cardiac Arrest Days Before It Strikes
AI can now predict deadly heart arrhythmias up to two weeks in advance, potentially transforming cardiac care. Artificial intelligence could play a key role in preventing many cases of [...]
NanoApps Medical is a Top 20 Feedspot Nanotech Blog
There is an ocean of Nanotechnology news published every day. Feedspot saves us a lot of time and we recommend it. We have been using it since 2018. [...]
This Startup Says It Can Clean Your Blood of Microplastics
This is a non-exhaustive list of places microplastics have been found: Mount Everest, the Mariana Trench, Antarctic snow, clouds, plankton, turtles, whales, cattle, birds, tap water, beer, salt, human placentas, [...]
New Blood Test Detects Alzheimer’s and Tracks Its Progression With 92% Accuracy
The new test could help identify which patients are most likely to benefit from new Alzheimer’s drugs. A newly developed blood test for Alzheimer’s disease not only helps confirm the presence [...]
The CDC buried a measles forecast that stressed the need for vaccinations
This story was originally published on ProPublica, a nonprofit newsroom that investigates abuses of power. Sign up to receive our biggest stories as soon as they’re published. ProPublica — Leaders at the Centers [...]
Light-Driven Plasmonic Microrobots for Nanoparticle Manipulation
A recent study published in Nature Communications presents a new microrobotic platform designed to improve the precision and versatility of nanoparticle manipulation using light. Led by Jin Qin and colleagues, the [...]
Cancer’s “Master Switch” Blocked for Good in Landmark Study
Researchers discovered peptides that permanently block a key cancer protein once thought untreatable, using a new screening method to test their effectiveness inside cells. For the first time, scientists [...]
AI self-cloning claims: A new frontier or a looming threat?
Chinese scientists claim that some AI models can replicate themselves and protect against shutdown. Has artificial intelligence crossed the so-called red line? Chinese researchers have published two reports on [...]
New Drug Turns Human Blood Into Mosquito-Killing Weapon
Nitisinone, a drug for rare diseases, kills mosquitoes when present in human blood and may become a new tool to fight malaria, offering longer-lasting, environmentally safer effects than ivermectin. [...]
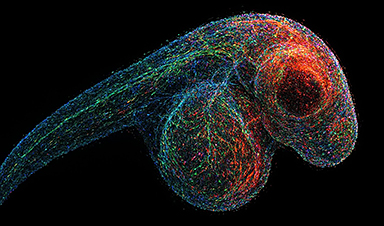
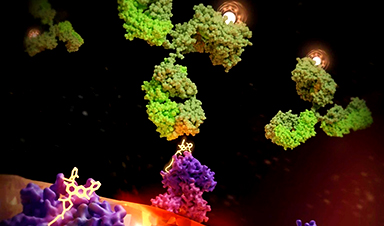
DNA Microscopy Creates 3D Maps of Life From the Inside Out
What if you could take a picture of every gene inside a living organism—not with light, but with DNA itself? Scientists at the University of Chicago have pioneered a revolutionary imaging technique called [...]
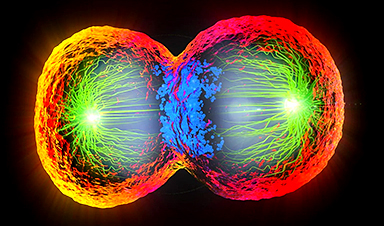
Scientists Just Captured the Stunning Process That Shapes Chromosomes
Scientists at EMBL have captured how human chromosomes fold into their signature rod shape during cell division, using a groundbreaking method called LoopTrace. By observing overlapping DNA loops forming in high [...]
Bird Flu Virus Is Mutating Fast – Scientists Say Our Vaccines May Not Be Enough
H5N1 influenza is evolving rapidly, weakening the effectiveness of existing antibodies and increasing its potential threat to humans. Scientists at UNC Charlotte and MIT used high-performance computational modeling to analyze thousands [...]
Revolutionary Cancer Vaccine Targets All Solid Tumors
The method triggers immune responses that inhibit melanoma, triple-negative breast cancer, lung carcinoma, and ovarian cancer. Cancer treatment vaccines have been in development since 2010, when the first was [...]
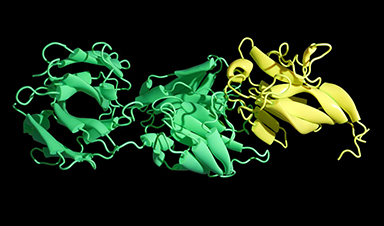
Scientists Uncover Hidden Protein Driving Autoimmune Attacks
Scientists have uncovered a critical piece of the puzzle in autoimmune diseases: a protein that helps release immune response molecules. By studying an ultra-rare condition, researchers identified ArfGAP2 as [...]
Mediterranean neutrino observatory sets new limits on quantum gravity
Quantum gravity is the missing link between general relativity and quantum mechanics, the yet-to-be-discovered key to a unified theory capable of explaining both the infinitely large and the infinitely [...]
Challenging Previous Beliefs: Japanese Scientists Discover Hidden Protector of Heart
A Japanese research team found that the oxidized form of glutathione (GSSG) may protect heart tissue by modifying a key protein, potentially offering a novel therapeutic approach for ischemic [...]
Millions May Have Long COVID – So Why Can’t They Get Diagnosed?
Millions of people in England may be living with Long Covid without even realizing it. A large-scale analysis found that nearly 10% suspect they might have the condition but [...]
Researchers Reveal What Happens to Your Brain When You Don’t Get Enough Sleep
What if poor sleep was doing more than just making you tired? Researchers have discovered that disrupted sleep in older adults interferes with the brain’s ability to clean out [...]
How to prevent chronic inflammation from zombie-like cells that accumulate with age
In humans and other multicellular organisms, cells multiply. This defining feature allows embryos to grow into adulthood, and enables the healing of the many bumps, bruises and scrapes along [...]
Breakthrough for long Covid patients who lost sense of smell
A breakthrough nasal surgery has restored the sense of smell for a dozen long Covid patients. Experts at University College London Hospitals NHS Foundation Trust successfully employed a technique typically used [...]
Scientists Invent Plastic That Can Dissolve In Seawater In Just A Few Hours
Plastic waste and pollution in the sea have been among the most serious environmental problems for decades, causing immense damage to marine life and ecosystems. However, a breakthrough discovery [...]
Muscles from the 3D printer
Swiss researchers have developed a method for printing artificial muscles out of silicone. In the future, these could be used on both humans and robots. Swiss researchers have succeeded [...]
Beneficial genetic changes observed in regular blood donors
Researchers at the Francis Crick Institute have identified genetic changes in blood stem cells from frequent blood donors that support the production of new, non-cancerous cells. Understanding the differences [...]
Shocking Amounts of Microplastics in the Brain – It Could Be Increasing Our Risk of Dementia
The brain has higher concentrations of plastic particles compared to other organs, with increased levels found in dementia patients. In a comprehensive commentary published in Brain Medicine, researchers highlight alarming [...]
Baffling Scientists for Centuries: New Study Unravels Mystery of Static Electricity
ISTA physicists demonstrate that contact electrification depends on the contact history of materials. For centuries, static electricity has intrigued and perplexed scientists. Now, researchers from the Waitukaitis group at [...]
Tumor “Stickiness” – Scientists Develop Potential New Way To Predict Cancer’s Spread
UC San Diego researchers have developed a device that predicts breast cancer aggressiveness by measuring tumor cell adhesion. Weakly adherent cells indicate a higher risk of metastasis, especially in [...]
Scientists Just Watched Atoms Move for the First Time Using AI
Scientists have developed a groundbreaking AI-driven technique that reveals the hidden movements of nanoparticles, essential in materials science, pharmaceuticals, and electronics. By integrating artificial intelligence with electron microscopy, researchers can now [...]
Scientists Sound Alarm: “Safe” Antibiotic Has Led to an Almost Untreatable Superbug
A recent study reveals that an antibiotic used for liver disease patients may increase their risk of contracting a dangerous superbug. An international team of researchers has discovered that [...]
Scientists Discover Natural Compound That Stops Cancer Progression
A discovery led by OHSU was made possible by years of study conducted by University of Portland undergraduates. Scientists have discovered a natural compound that can halt a key [...]
Scientists Just Discovered an RNA That Repairs DNA Damage – And It’s a Game-Changer
Our DNA is constantly under threat — from cell division errors to external factors like sunlight and smoking. Fortunately, cells have intricate repair mechanisms to counteract this damage. Scientists have uncovered [...]
What Scientists Just Discovered About COVID-19’s Hidden Death Toll
COVID-19 didn’t just claim lives directly—it reshaped mortality patterns worldwide. A major international study found that life expectancy plummeted across most of the 24 analyzed countries, with additional deaths from cardiovascular [...]
Self-Propelled Nanoparticles Improve Immunotherapy for Non-Invasive Bladder Cancer
A study led by Pohang University of Science and Technology (POSTECH) and the Institute for Bioengineering of Catalonia (IBEC) in South Korea details the creation of urea-powered nanomotors that enhance immunotherapy [...]
Scientists Develop New System That Produces Drinking Water From Thin Air
UT Austin researchers have developed a biodegradable, biomass-based hydrogel that efficiently extracts drinkable water from the air, offering a scalable, sustainable solution for water access in off-grid communities, emergency [...]
AI Unveils Hidden Nanoparticles – A Breakthrough in Early Disease Detection
Deep Nanometry (DNM) is an innovative technique combining high-speed optical detection with AI-driven noise reduction, allowing researchers to find rare nanoparticles like extracellular vesicles (EVs). Since EVs play a [...]
Inhalable nanoparticles could help treat chronic lung disease
Nanoparticles designed to release antibiotics deep inside the lungs reduced inflammation and improved lung function in mice with symptoms of chronic obstructive pulmonary disease By Grace Wade Delivering medication to [...]
New MRI Study Uncovers Hidden Lung Abnormalities in Children With Long COVID
Long COVID is more than just lingering symptoms—it may have a hidden biological basis that standard medical tests fail to detect. A groundbreaking study using advanced MRI technology has [...]
AI Struggles with Abstract Thought: Study Reveals GPT-4’s Limits
While GPT-4 performs well in structured reasoning tasks, a new study shows that its ability to adapt to variations is weak—suggesting AI still lacks true abstract understanding and flexibility [...]
Turning Off Nerve Signals: Scientists Develop Promising New Pancreatic Cancer Treatment
Pancreatic cancer reprograms nerve cells to fuel its growth, but blocking these connections can shrink tumors and boost treatment effectiveness. Pancreatic cancer is closely linked to the nervous system, [...]
New human antibody shows promise for Ebola virus treatment
New research led by scientists at La Jolla Institute for Immunology (LJI) reveals the workings of a human antibody called mAb 3A6, which may prove to be an important [...]
Early Alzheimer’s Detection Test – Years Before Symptoms Appear
A new biomarker test can detect early-stage tau protein clumping up to a decade before it appears on brain scans, improving early Alzheimer’s diagnosis. Unlike amyloid-beta, tau neurofibrillary tangles are directly [...]
New mpox variant can spread rapidly across borders
International researchers, including from DTU National Food Institute, warn that the ongoing mpox outbreak in the Democratic Republic of the Congo (DRC) has the potential to spread across borders [...]
How far would you trust AI to make important decisions?
From tailored Netflix recommendations to personalized Facebook feeds, artificial intelligence (AI) adeptly serves content that matches our preferences and past behaviors. But while a restaurant tip or two is [...]
Can AI Really Think? Research Reveals Gaps in Logical Execution
While AI models can break down problems into structured steps, new research reveals they still fail at basic arithmetic and fact-checking—raising questions about their true reasoning abilities. Large Language [...]
Scientists Just Made Cancer Radiation Therapy Smarter, Safer, and More Precise
Scientists at UC San Francisco have developed a revolutionary cancer treatment that precisely targets tumors with radiation while sparing healthy tissues. By using a KRAS-targeting drug to mark cancer [...]
Superbugs Are Losing to Science, Light, and a Little Spice
Texas A&M researchers have found that curcumin, when activated by light, can weaken antibiotic-resistant bacteria, restoring the effectiveness of conventional antibiotics. Curcumin: A Surprising Ally Against Superbugs In 2017, [...]
New Research Shatters the Perfect Pitch Myth
For decades, people believed absolute pitch was an exclusive ability granted only to those with the right genetics or early music training. But new research from the University of [...]
Why Some Drinkers Suffer Devastating Liver Damage While Others Don’t
A study from Keck Medicine of USC found that heavy drinkers with diabetes, high blood pressure, or a large waistline are up to 2.4 times more likely to develop advanced liver [...]
“Good” Cholesterol Could Be Bad for Your Eyes – New Study Raises Concerns
‘Good’ cholesterol may be linked to an increased risk of glaucoma in individuals over 55, while, paradoxically, ‘bad’ cholesterol may be associated with a lower risk. These findings challenge [...]
Reawakening Dormant Nerve Cells: Groundbreaking Neurotechnology Restores Motor Function
A new electrical stimulation therapy for spinal muscle atrophy (SMA) has shown promise in reactivating motor neurons and improving movement. In a pilot clinical trial, three patients who received [...]




























































Leave A Comment