A study recently published in the journal Infection, Genetics and Evolution explored the attachment affinity of carbon nanotubes (CNTs) and carbon nano-fullerene towards numerous molecular targets of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Computational modeling of the 3D architectures of nano-fullerenes and CNTs was carried out, and molecule binding and molecular dynamic (MD) simulations were used to estimate the attachment affinity of the nanoparticles to the chosen target molecules. The research emphasizes the need of using carbon nanoparticles as a treatment for COVID-19.
COVID-19 Taking the World by Storm
The shocking COVID-19 epidemic attributed to Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), first detected in Wuhan, China, in December of 2019, has since expanded globally. A virulent pathogen, SARS-CoV-2 belongs to the ß-coronavirus family and is a positive-stranded RNA virus. The spike glycoproteins that appear on the envelope give these pathogens a crown-like appearance. Other proteins in the viral structure include membrane and envelope proteins, nucleocapsid protein, and RNA dependent on RNA polymerase as structural proteins.
Inadequacy of Current Treatments
At first, anti-viral medications including remdesivir, lopinavir, chloroquine, and hydroxychloroquine were recommended for COVID-19 infection therapy. Recent assessments, however, proved the ineffectiveness of chloroquine and hydroxychloroquine when combatting SARS-CoV-2. Several drugs are in the early phases of clinical development.
Novel variants of the virus have been discovered in various regions across the globe, with these mutations exhibiting enhanced transmission and pathogenicity, as well as reduced neutralization.
Vaccines are now available to treat COVID-19, although they have not yet reached populations in some countries. The variety of symptoms found in the patients, as well as asymptomatic transmission and vaccination resistance data, indicate that the creation of an alternative treatment answer is imminently needed.
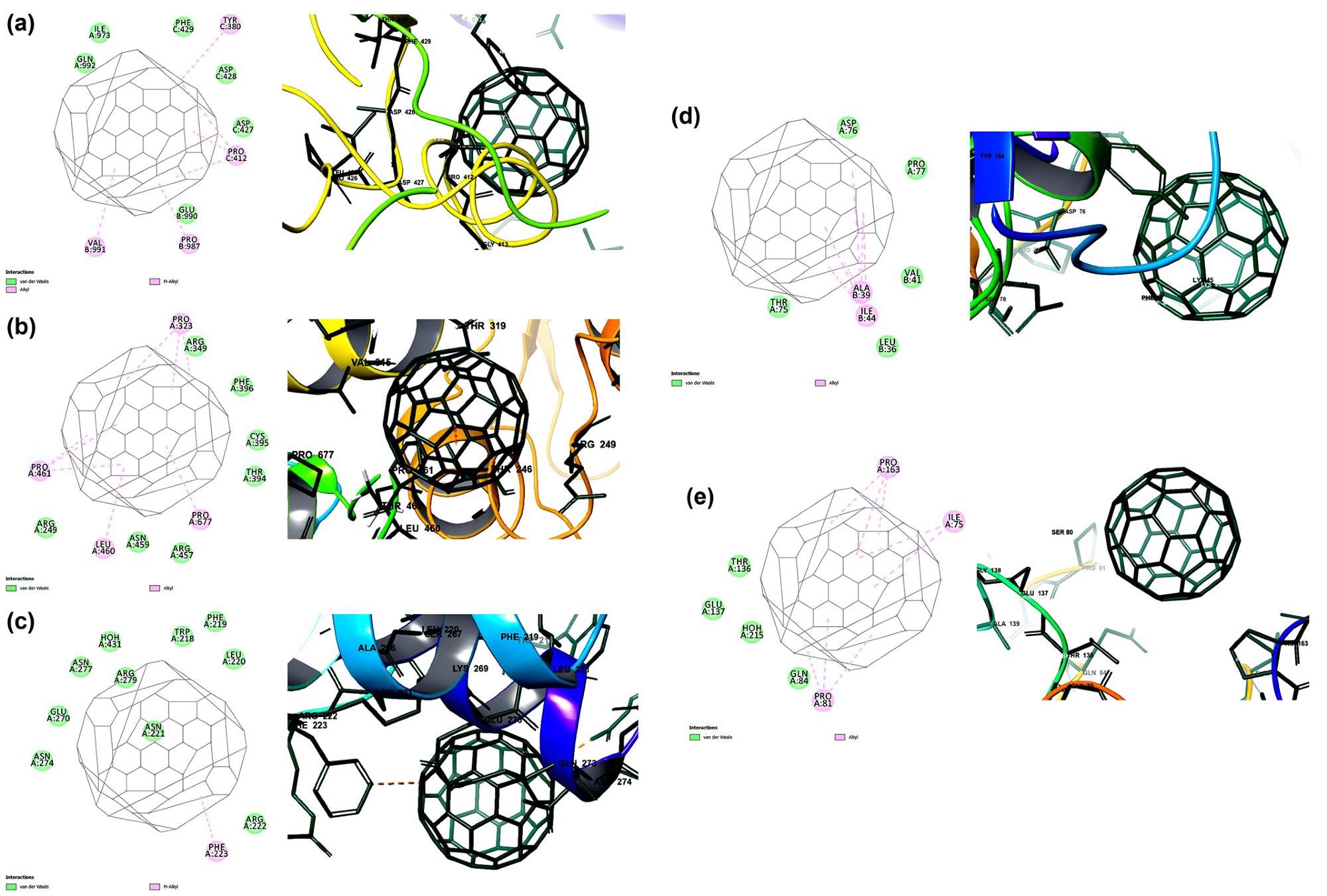
The binding potential of carbon nanofullerene towards the prioritized targets of SARS-CoV-2 is predicted by molecular docking. The binding affinity and interactions of carbon nano fullerene towards (a) spike glycoprotein (−13.7 kcal/mol), (b) RNA dependent RNA polymerase (−12.9 kcal/mol), (c) main protease (−11.4 kcal/mol), (d) papain-like protease (−10.6 kcal/mol) and (d) RNA binding domain of nucleocapsid protein (−10.1 kcal/mol). Image Credit: Skariyachan, S. et al
Nanotechnology and Computational Biology
Modern developments in nanotechnology have revealed the possibility of using nanoparticles such as CNTs and nano-fullerenes to target various areas of SARS-CoV-2, blocking its pathogenic effects. Carbon nanotubes are being touted as potential therapeutic materials owing to excellent mechanical capabilities, structural soundness, and tunability of functional groups.
Nanomaterials may be used to create nano-based COVID-19 protective devices and disinfectants. Furthermore, nanoparticles could be employed to function as antigen carriers or serve as an adjuvant medication for use concurrently with the upcoming COVID-19 vaccines.
The team used computation-based virtual screening and MD simulations to determine the binding capability of carbon nanotubes and nano-fullerenes to numerous putative target areas of the virus. Carbon nano-fullerenes have a potential affinity for SARS-CoV-2 targets, and carbon nanotubes have demonstrated possible interactions that might limit the virus’s harmful mechanism.
Identifying Structural Properties of Recognized Targets
Spike glycoproteins are the principal target of antibodies and are important in stimulating entrance into cells through the transmembrane spike. The transmembrane spike is composed of two functional subunits that are in charge of attaching to receptors of host cells and fusing the membranes of the virus and cells.
Each virus may identify distinct places of attachment and entrance by connecting with specific areas of the receptors in the host cell unit, depending on the type of viral strain. The major constituent of the viral machinery is responsible for replication and transcription; the RNA-dependent RNA polymerase (RdRp) is required for the survival of these viruses. Treatments targeting this area would be an excellent strategy.
The primary protease is an enzyme that is also involved in viral replication and transcription. The major protease is required for the digestion of viral RNA-translated polyproteins. Inhibiting this enzyme may aid in the prevention of viral replication. As a result, these proteins were chosen in the study because they might be potential therapeutic targets for SARS-CoV-2 infection.
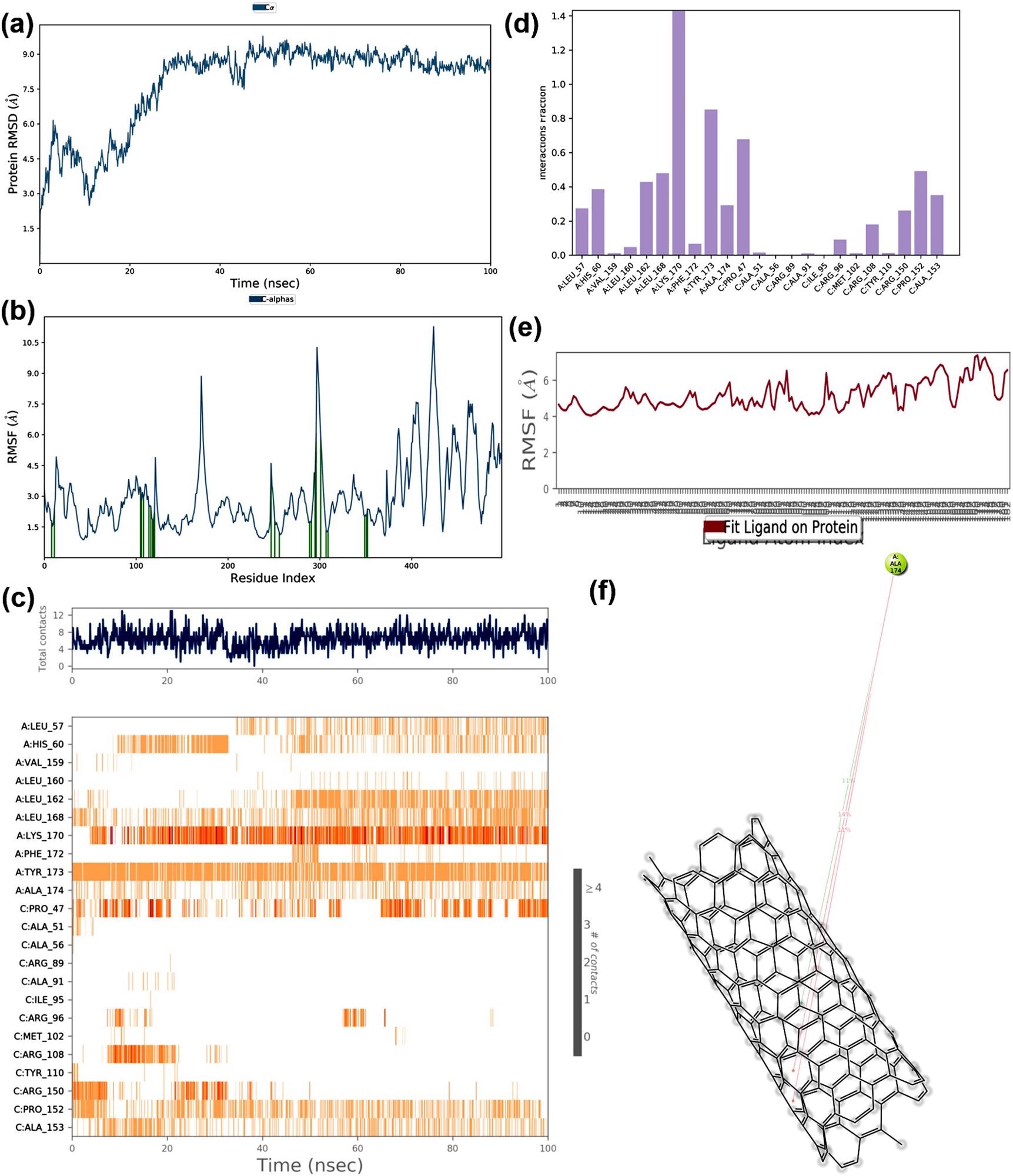
The MD simulation trajectories of RNA binding domain of the nucleocapsid protein and nano tube complex (a) RMSD: Protein RMSD (Å) on the y-axis and time on the x-axis (b) Protein RMSF (Å) on the y-axis and residues on the x-axis (c) Protein-ligand contacts over the simulation course, (d) histogram representing interaction fraction on the y-axis and residues on the x-axis (e) Ligand RMSF (Å) on the y-axis and atoms on the x-axis and (f) Major interactions that occur during MD simulation. Image Credit: Skariyachan, S. et al
Conclusions
It may initially seem unimpressive that the targets adopt this orientation only after adsorption on hydrophobic surfaces. However, there is potential for various orientations concerning the adsorption process on hydrophobic or charged areas. Therefore, understanding the interaction modeling of many orientations is critical.
The present research lays the groundwork for future laboratory tests and exploratory trials. Despite the nanoparticles’ apparent toxic nature, they might be employed as possible leads for blocking the coronavirus target areas. Research has shown the success of their use in target-specified treatments, drug transportation processes, cancer-related therapies, and other applications.
The MD simulation tests indicated that the priority entities and nanoparticles undergo dynamic interactions, which are steady and promising. When compared to carbon-fullerenes, nanotubes have the higher binding energy of the two carbon nanoparticles.
News
New Once-a-Week Shot Promises Life-Changing Relief for Parkinson’s Patients
A once-a-week shot from Australian scientists could spare people with Parkinson’s the grind of taking pills several times a day. The tiny, biodegradable gel sits under the skin and releases steady doses of two [...]
Weekly injectable drug offers hope for Parkinson’s patients
A new weekly injectable drug could transform the lives of more than eight million people living with Parkinson's disease, potentially replacing the need for multiple daily tablets. Scientists from the University of South Australia [...]
Most Plastic in the Ocean Is Invisible—And Deadly
Nanoplastics—particles smaller than a human hair—can pass through cell walls and enter the food web. New research suggest 27 million metric tons of nanoplastics are spread across just the top layer of the North [...]
Repurposed drugs could calm the immune system’s response to nanomedicine
An international study led by researchers at the University of Colorado Anschutz Medical Campus has identified a promising strategy to enhance the safety of nanomedicines, advanced therapies often used in cancer and vaccine treatments, [...]
Nano-Enhanced Hydrogel Strategies for Cartilage Repair
A recent article in Engineering describes the development of a protein-based nanocomposite hydrogel designed to deliver two therapeutic agents—dexamethasone (Dex) and kartogenin (KGN)—to support cartilage repair. The hydrogel is engineered to modulate immune responses and promote [...]
New Cancer Drug Blocks Tumors Without Debilitating Side Effects
A new drug targets RAS-PI3Kα pathways without harmful side effects. It was developed using high-performance computing and AI. A new cancer drug candidate, developed through a collaboration between Lawrence Livermore National Laboratory (LLNL), BridgeBio Oncology [...]
Scientists Are Pretty Close to Replicating the First Thing That Ever Lived
For 400 million years, a leading hypothesis claims, Earth was an “RNA World,” meaning that life must’ve first replicated from RNA before the arrival of proteins and DNA. Unfortunately, scientists have failed to find [...]
Why ‘Peniaphobia’ Is Exploding Among Young People (And Why We Should Be Concerned)
An insidious illness is taking hold among a growing proportion of young people. Little known to the general public, peniaphobia—the fear of becoming poor—is gaining ground among teens and young adults. Discover the causes [...]
Team finds flawed data in recent study relevant to coronavirus antiviral development
The COVID pandemic illustrated how urgently we need antiviral medications capable of treating coronavirus infections. To aid this effort, researchers quickly homed in on part of SARS-CoV-2's molecular structure known as the NiRAN domain—an [...]
Drug-Coated Neural Implants Reduce Immune Rejection
Summary: A new study shows that coating neural prosthetic implants with the anti-inflammatory drug dexamethasone helps reduce the body’s immune response and scar tissue formation. This strategy enhances the long-term performance and stability of electrodes [...]
Scientists discover cancer-fighting bacteria that ‘soak up’ forever chemicals in the body
A family of healthy bacteria may help 'soak up' toxic forever chemicals in the body, warding off their cancerous effects. Forever chemicals, also known as PFAS (per- and polyfluoroalkyl substances), are toxic chemicals that [...]
Johns Hopkins Researchers Uncover a New Way To Kill Cancer Cells
A new study reveals that blocking ribosomal RNA production rewires cancer cell behavior and could help treat genetically unstable tumors. Researchers at the Johns Hopkins Kimmel Cancer Center and the Department of Radiation Oncology and Molecular [...]
AI matches doctors in mapping lung tumors for radiation therapy
In radiation therapy, precision can save lives. Oncologists must carefully map the size and location of a tumor before delivering high-dose radiation to destroy cancer cells while sparing healthy tissue. But this process, called [...]
Scientists Finally “See” Key Protein That Controls Inflammation
Researchers used advanced microscopy to uncover important protein structures. For the first time, two important protein structures in the human body are being visualized, thanks in part to cutting-edge technology at the University of [...]
AI tool detects 9 types of dementia from a single brain scan
Mayo Clinic researchers have developed a new artificial intelligence (AI) tool that helps clinicians identify brain activity patterns linked to nine types of dementia, including Alzheimer's disease, using a single, widely available scan—a transformative [...]
Is plastic packaging putting more than just food on your plate?
New research reveals that common food packaging and utensils can shed microscopic plastics into our food, prompting urgent calls for stricter testing and updated regulations to protect public health. Beyond microplastics: The analysis intentionally [...]