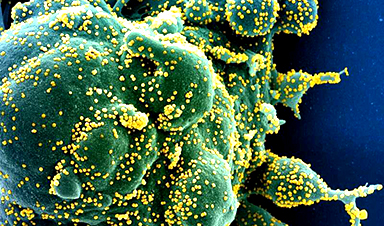
The COVID-19 pandemic has prompted considerable investigation into how the SARS-CoV-2 Spike protein attaches to a human cell during the infection process, as this knowledge is useful in designing vaccines and therapeutics. Now, a team of scientists has discovered additional locations on the Spike protein that may not only help to explain how certain mutations make emerging variants more infectious but also could be used as additional targets for therapeutic intervention.
“Significant research is underway to examine how the receptor binding domain (RBD) at the tip of the club-shaped SARS-CoV-2 Spike protein attaches to an ACE2 receptor on a human cell, but little is known about the other changes that occur in the Spike protein as a result of this attachment,” said Ganesh Anand, associate professor of chemistry, Penn State. “We have uncovered ‘hotspots’ further down on the Spike protein that are critical for SARS-CoV-2 infection and may be novel targets beyond the RBD for therapeutic intervention.”
Anand and his colleagues used a process, called amide hydrogen-deuterium exchange mass spectrometry (HDXMS), to visualize what happens when the SARS-CoV-2 Spike protein binds to an ACE2 receptor. HDXMS uses heavy water or deuterium oxide (D2O), a naturally occurring, non-radioactive isotope of water formed from heavy hydrogen or deuterium, as a probe for mapping proteins. In this case, the team placed SARS-CoV-2 Spike protein and ACE2 receptors in heavy water and obtained footprints of ACE2 on the Spike protein.
“If you put the Spike protein and ACE2 receptor into a solution that’s made with D2O, the surfaces and more floppy regions on both proteins will more readily exchange hydrogens for deuterium, compared to their interiors,” said Anand. “And footprints of each protein on the binding partner can be readily identified from areas where you see little deuterium and only detect normal hydrogen.”
Using this technique, the team determined that binding of the Spike protein and ACE2 receptor is necessary for furin-like proteases—a family of human enzymes—that act to snip off the tip, called the S1 subunit, of the Spike protein, which is the next step in the virus’s infection of the cell. The findings published on Feb. 8 in the journal eLife.
Image Credit: NIH/NIAID
Post by Amanda Scott, NA CEO. Follow her on twitter @tantriclens
Thanks to Heinz V. Hoenen. Follow him on twitter: @HeinzVHoenen
News
Repurposed drugs could calm the immune system’s response to nanomedicine
An international study led by researchers at the University of Colorado Anschutz Medical Campus has identified a promising strategy to enhance the safety of nanomedicines, advanced therapies often used in cancer and vaccine treatments, [...]
Nano-Enhanced Hydrogel Strategies for Cartilage Repair
A recent article in Engineering describes the development of a protein-based nanocomposite hydrogel designed to deliver two therapeutic agents—dexamethasone (Dex) and kartogenin (KGN)—to support cartilage repair. The hydrogel is engineered to modulate immune responses and promote [...]
New Cancer Drug Blocks Tumors Without Debilitating Side Effects
A new drug targets RAS-PI3Kα pathways without harmful side effects. It was developed using high-performance computing and AI. A new cancer drug candidate, developed through a collaboration between Lawrence Livermore National Laboratory (LLNL), BridgeBio Oncology [...]
Scientists Are Pretty Close to Replicating the First Thing That Ever Lived
For 400 million years, a leading hypothesis claims, Earth was an “RNA World,” meaning that life must’ve first replicated from RNA before the arrival of proteins and DNA. Unfortunately, scientists have failed to find [...]
Why ‘Peniaphobia’ Is Exploding Among Young People (And Why We Should Be Concerned)
An insidious illness is taking hold among a growing proportion of young people. Little known to the general public, peniaphobia—the fear of becoming poor—is gaining ground among teens and young adults. Discover the causes [...]
Team finds flawed data in recent study relevant to coronavirus antiviral development
The COVID pandemic illustrated how urgently we need antiviral medications capable of treating coronavirus infections. To aid this effort, researchers quickly homed in on part of SARS-CoV-2's molecular structure known as the NiRAN domain—an [...]
Drug-Coated Neural Implants Reduce Immune Rejection
Summary: A new study shows that coating neural prosthetic implants with the anti-inflammatory drug dexamethasone helps reduce the body’s immune response and scar tissue formation. This strategy enhances the long-term performance and stability of electrodes [...]
Scientists discover cancer-fighting bacteria that ‘soak up’ forever chemicals in the body
A family of healthy bacteria may help 'soak up' toxic forever chemicals in the body, warding off their cancerous effects. Forever chemicals, also known as PFAS (per- and polyfluoroalkyl substances), are toxic chemicals that [...]
Johns Hopkins Researchers Uncover a New Way To Kill Cancer Cells
A new study reveals that blocking ribosomal RNA production rewires cancer cell behavior and could help treat genetically unstable tumors. Researchers at the Johns Hopkins Kimmel Cancer Center and the Department of Radiation Oncology and Molecular [...]
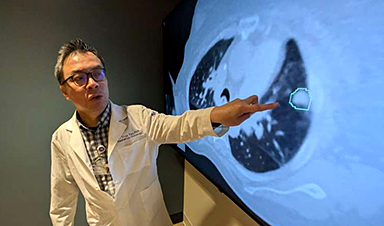
AI matches doctors in mapping lung tumors for radiation therapy
In radiation therapy, precision can save lives. Oncologists must carefully map the size and location of a tumor before delivering high-dose radiation to destroy cancer cells while sparing healthy tissue. But this process, called [...]
Scientists Finally “See” Key Protein That Controls Inflammation
Researchers used advanced microscopy to uncover important protein structures. For the first time, two important protein structures in the human body are being visualized, thanks in part to cutting-edge technology at the University of [...]
AI tool detects 9 types of dementia from a single brain scan
Mayo Clinic researchers have developed a new artificial intelligence (AI) tool that helps clinicians identify brain activity patterns linked to nine types of dementia, including Alzheimer's disease, using a single, widely available scan—a transformative [...]
Is plastic packaging putting more than just food on your plate?
New research reveals that common food packaging and utensils can shed microscopic plastics into our food, prompting urgent calls for stricter testing and updated regulations to protect public health. Beyond microplastics: The analysis intentionally [...]
Aging Spreads Through the Bloodstream
Summary: New research reveals that aging isn’t just a local cellular process—it can spread throughout the body via the bloodstream. A redox-sensitive protein called ReHMGB1, secreted by senescent cells, was found to trigger aging features [...]
AI and nanomedicine find rare biomarkers for prostrate cancer and atherosclerosis
Imagine a stadium packed with 75,000 fans, all wearing green and white jerseys—except one person in a solid green shirt. Finding that person would be tough. That's how hard it is for scientists to [...]
Are Pesticides Breeding the Next Pandemic? Experts Warn of Fungal Superbugs
Fungicides used in agriculture have been linked to an increase in resistance to antifungal drugs in both humans and animals. Fungal infections are on the rise, and two UC Davis infectious disease experts, Dr. George Thompson [...]