The bonding interaction amongst Influenza A and the peptide “PeB,” which selectively binds the viral surface protein hemagglutinin, has been investigated using electrically controlled deoxyribonuclic acid (DNA) nanolevers in the journal Advanced Materials.
PeB is conjugated to DNA strands that are bonded to complementary anchors and fixed on the electrode surface of a “switchSENSE” biochip. A fluorophore is attached to the surface-tethered DNA strand, whereas the complementary strand has a multivalent configuration containing up to three PeB peptides. A negative voltage is used to keep the nanolevers erect (static).
As the current epidemic of the SARS-CoV-2 virus has demonstrated, viral pandemics represent a tremendous threat to humanity. A similar pandemic, known as the Spanish flu, was induced by an influenza A virus and led to millions of fatalities around the world. Annual outbreaks of different intensity are still caused by influenza A viruses. The globe was recently confronted with the swine flu in 2009.
Influenza A is an Orthomyxoviridae virus that is encased in a lipid bilayer that contains three cell membranes: hemagglutinin (HA), neuraminidase (NA), and the M2 proton channels. Neuraminidase is a glycoside cleaver that is essential for the discharge of viral particles from infected cells.
Subsequent to viral fusing, hemagglutinin binds to the host cell’s sialic acid (SA)-containing cellular receptors. Understanding viral internalization and infection requires kinetic analysis of affinity and fervency coefficients of various receptor binding to hemagglutinin.
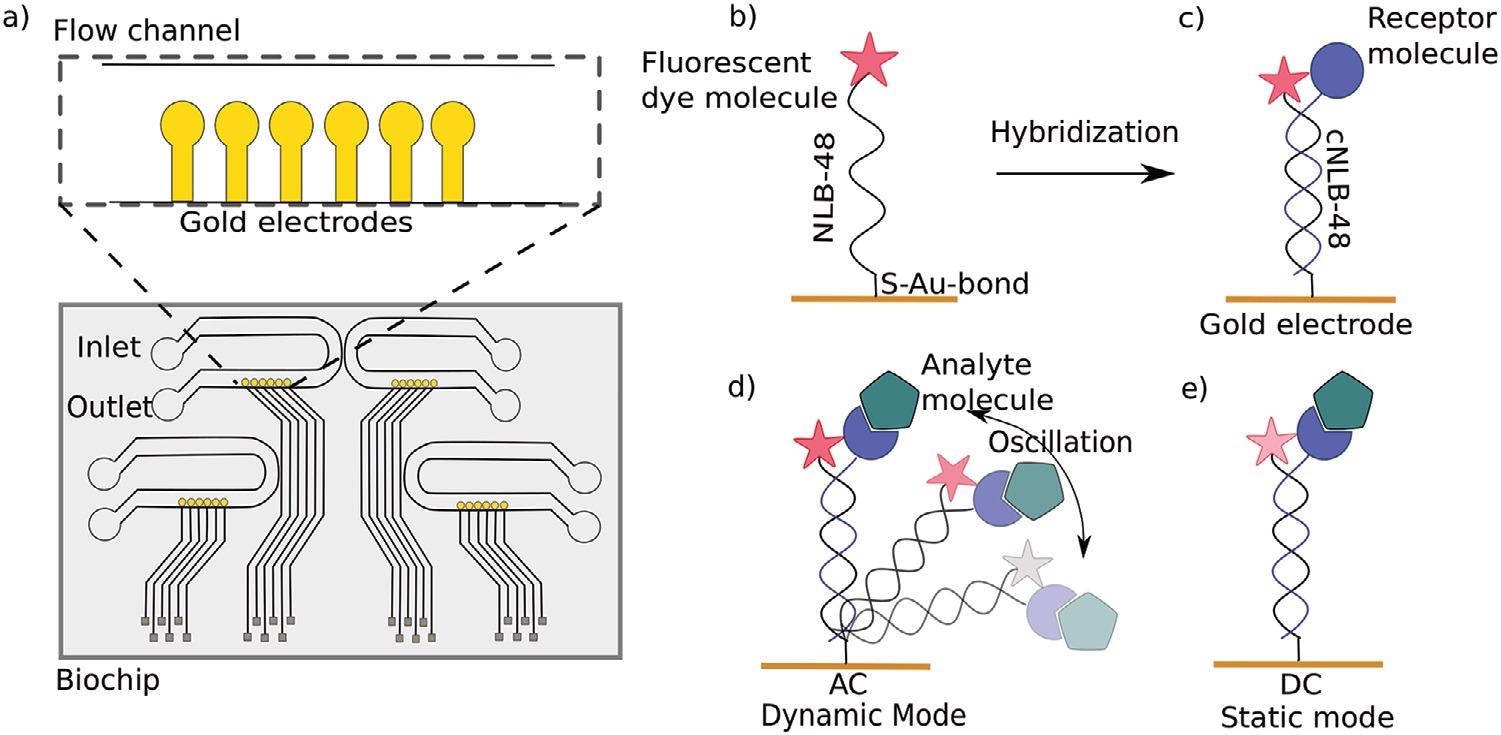
a) Top view of a switchSENSE microfluidic biochip with four flow channels. Each channel comprises six gold electrodes. The more detailed view shows the six independent measurement electrodes in a row within the microfluidic channel. b) Schematic overview of a single stranded DNA nanolever (NL-B48) immobilized on a gold electrode via thiol coupling. The nanolever carries a fluorophore at the lateral end. c) The DNA monolayer is functionalized with the ligand of interest by hybridization of the complementary DNA strand carrying a receptor molecule at the distal end. d) Dynamic measurement mode: The double stranded DNA nanolever is actuated to a switching motion by applying an alternating potential. The fluorescence signal is gradually quenched by energy transfer upon approaching the gold surface. Read-out of this mode is the switching speed of DNA nanolevers. The switching speed is slowed down upon binding of an analyte to the ligand molecule adding friction to the nanolever. e) Static measurement mode: DNA nanolevers are kept at an upright position. Read-out of this mode is the change in fluorescence intensity upon binding of an analyte due to changes in the local environment of the dye. Drawings are not to scale. Image Credit: Kruse M et al., Advanced Material
The ability to calculate not just the dissociation constant but also rate constants is one of the key features of the switchSENSE technique. These kinetic metrics are becoming increasingly important in determining the effectiveness of candidates in medication development.
Other methods for determining dissociation constants exist, but they lack the capacity to resolve bonding kinetic parameters. Radioligand binding tests, microscale thermophoresis (MST), and isothermal calorimetry are some examples of these.
Surface plasmon resonance is one technology that allows for real-time monitoring of kinetic parameters (SPR). SPR measures and switchSENSE technologies are similar in that they both have benefits and drawbacks. As a result, a similar naming system is used.
The quantifiable metrics of kinetic parameters in a timely manner, label-free detection, minimal sample expenditure, and high sensitivity of the technology are all benefits of SPR and switchSENSE. The disadvantages of both procedures are that one of the participants must be immobilized, which may have an impact on binding behavior, and that mass transfer is limited….
News
Completely New Use Discovered – This Traditional Herb Has Remarkable Nerve Regenerative Properties
Blessed thistle (Cnicus benedictus), a member of the Asteraceae family, thrives in our climate. This plant has been utilized for centuries as a medicinal herb, often consumed as an extract or tea to support [...]
Scientists study lipids cell by cell, making new cancer research possible
Imagine being able to look inside a single cancer cell and see how it communicates with its neighbors. Scientists are celebrating a new technique that lets them study the fatty contents of cancer cells, [...]
Antibiotic Breakthrough: Revolutionary Chinese Study Paves Way for Superbug Defeating Drugs
New research reveals that fluorous lipopetides act as highly effective antibiotics. Bacterial infections resistant to multiple drugs, which no existing antibiotics can treat, represent a significant worldwide challenge. A research group from China has [...]
Signs of Multiple Sclerosis Show Up in Blood Years Before Symptoms Appear
UCSF scientists clear a potential path toward earlier treatment for a disease that affects nearly 1,000,000 people in the United States. By Levi Gadye In a discovery that could hasten treatment for patients with multiple [...]
Advanced RNA Sequencing Reveals the Drivers of New COVID Variants
A study reveals that a new sequencing technique, tARC-seq, can accurately track mutations in SARS-CoV-2, providing insights into the rapid evolution and variant development of the virus. The SARS-CoV-2 virus that causes COVID has the unsettling [...]
No More Endless Boosters? Scientists Develop One-for-All Virus Vaccine
End of the line for endless boosters? Researchers at UC Riverside have developed a new vaccine approach using RNA that is effective against any strain of a virus and can be used safely even by babies or the immunocompromised. Every [...]
How Are Hydrogels Shaping the Future of Biomedicine?
Hydrogels have gained widespread recognition and utilization in biomedical engineering, with their applications dating back to the 1960s when they were first used in contact lens production. Hydrogels are distinguished from other biomaterials in [...]
Nanovials method for immune cell screening uncovers receptors that target prostate cancer
A recent UCLA study demonstrates a new process for screening T cells, part of the body's natural defenses, for characteristics vital to the success of cell-based treatments. The method filters T cells based on [...]
New Research Reveals That Your Sense of Smell May Be Smarter Than You Think
A new study published in the Journal of Neuroscience indicates that the sense of smell is significantly influenced by cues from other senses, whereas the senses of sight and hearing are much less affected. A popular [...]
Deadly bacteria show thirst for human blood: the phenomenon of bacterial vampirism
Some of the world's deadliest bacteria seek out and feed on human blood, a newly-discovered phenomenon researchers are calling "bacterial vampirism." A team led by Washington State University researchers has found the bacteria are [...]
Organ Architects: The Remarkable Cells Shaping Our Development
Finding your way through the winding streets of certain cities can be a real challenge without a map. To orient ourselves, we rely on a variety of information, including digital maps on our phones, [...]
Novel hydrogel removes microplastics from water
Microplastics pose a great threat to human health. These tiny plastic debris can enter our bodies through the water we drink and increase the risk of illnesses. They are also an environmental hazard; found [...]
Researchers Discover New Origin of Deep Brain Waves
Understanding hippocampal activity could improve sleep and cognition therapies. Researchers from the University of California, Irvine’s biomedical engineering department have discovered a new origin for two essential brain waves—slow waves and sleep spindles—that are critical for [...]
The Lifelong Cost of Surviving COVID: Scientists Uncover Long-Term Effects
Many of the individuals released to long-term acute care facilities suffered from conditions that lasted for over a year. Researchers at UC San Francisco studied COVID-19 patients in the United States who survived some of the longest and [...]
Previously Unknown Rogue Immune Key to Chronic Viral Infections Discovered
Scientists discovered a previously unidentified rogue immune cell linked to poor antibody responses in chronic viral infections. Australian researchers have discovered a previously unknown rogue immune cell that can cause poor antibody responses in [...]
Nature’s Betrayal: Unmasking Lead Lurking in Herbal Medicine
A case of lead poisoning due to Ayurvedic medicine use demonstrates the importance of patient history in diagnosis and the need for public health collaboration to prevent similar risks. An article in CMAJ (Canadian Medical Association [...]